T‐ALL Epigenetic Reprogramming Project
T-ALL, CRISPR-dCas9-TET1, epigenetic editing, TET2 reactivation
Lab
Prof. Colm Nestor’s group, Linköping University (LiU) – 2024
Focus
Using CRISPR-dCas9-TET1 to reactivate hypermethylated TET2 in T-ALL cells.
Overview
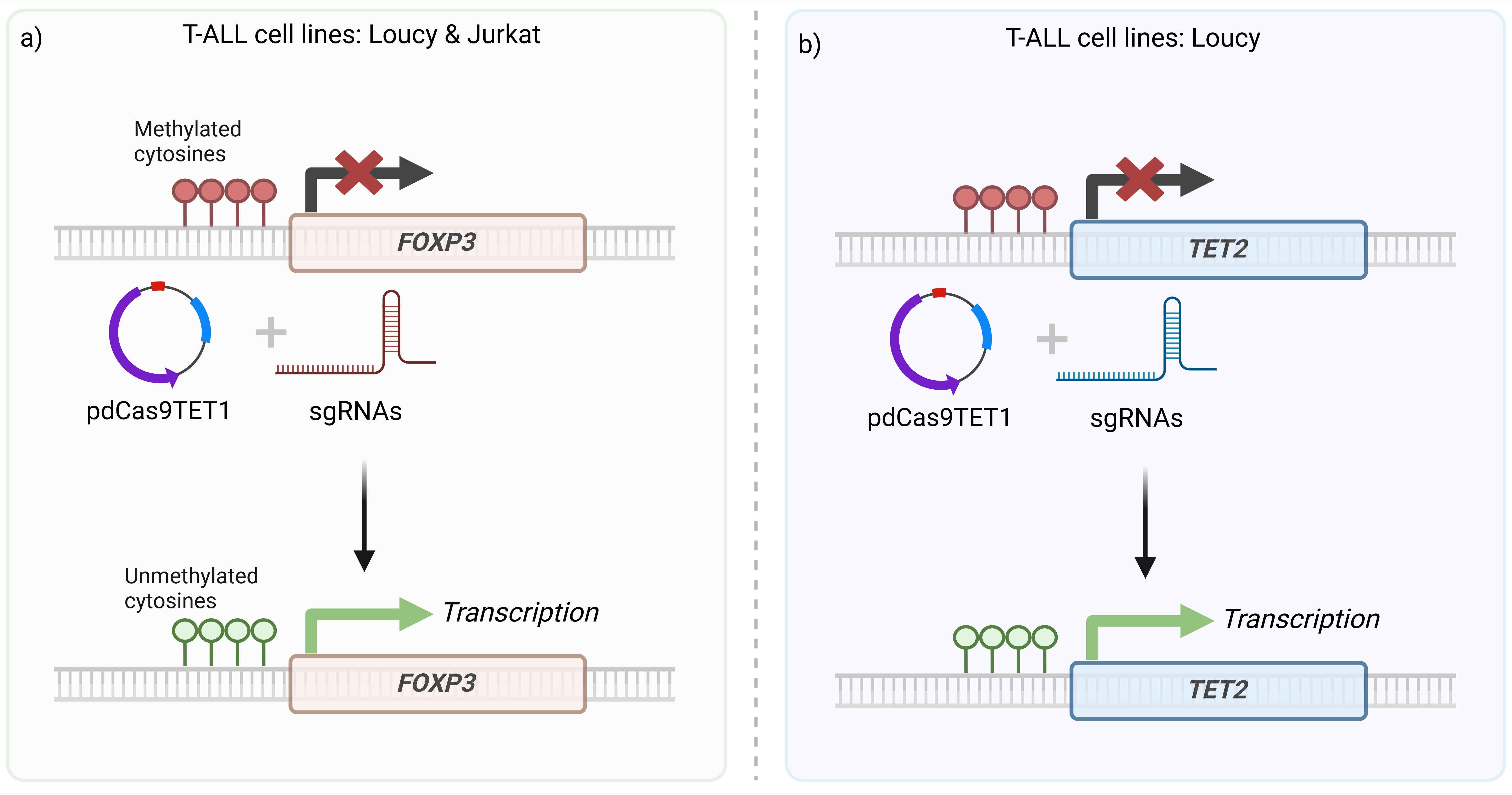
T-cell acute lymphoblastic leukemia (T-ALL) is a hematologic malignancy where tumor suppressor genes such as TET2 are frequently silenced due to promoter hypermethylation, disrupting normal cell regulation and contributing to leukemogenesis. TET2, part of the ten-eleven translocation (TET) enzyme family, plays a crucial role in DNA demethylation, which is essential for gene activation. Reversing this methylation may restore TET2 function, offering a potential therapeutic strategy.
Methods & Key Findings
Inspired by recent epigenetic editing studies on FOXP3 and CLTA, I applied and optimized these methodologies to target TET2. I designed sgRNAs for the TET2 promoter region, incorporated them into gBlocks, and verified their sequences through Sanger sequencing. The plasmid constructs were validated through restriction digestion and sequencing prior to transfection.
Transfection Optimization
Jurkat & Loucy Cell Transfection: Nucleofection was optimized to enhance transfection efficiency in suspension cells.
HEK293T Control Experiments: Lipofectamine transfection in HEK293T cells was used to confirm plasmid expression efficiency in adherent cells.
Assessment of Transfection Efficiency & Gene Reactivation:
- Fluorescence microscopy & flow cytometry assessed transfection success across all cell lines, using TagBFP as a marker.
- RT-qPCR analysis confirmed partial FOXP3 reactivation in Jurkat cells, validating CRISPRa-based epigenetic editing.
Key Takeaways
These results highlight the potential of epigenetic reprogramming as a therapeutic strategy in leukemia. Further optimizations are required to improve transfection efficiency and enhance gene reactivation outcomes.