Cardiomyocyte Differentiation
cardiomyocyte differentiation, hESC, Wnt signaling
Lab
Prof. Claudio Cantù’s group, Linköping University (LiU) – 2025
Focus
Investigating hESC differentiation into cardiomyocytes, emphasizing Wnt/β‐catenin signaling regulation.
Overview
This project aims to investigate dynamic epigenetic and chromatin accessibility changes during cardiomyocyte differentiation using a time-resolved approach. By integrating ATAC-seq, RNA-seq, and protein interaction assays, the study will map dynamic regulatory changes over differentiation stages, This will help uncover key Wnt signaling regulators, potentially revealing novel therapeutic targets for congenital heart disease.
Ongoing work
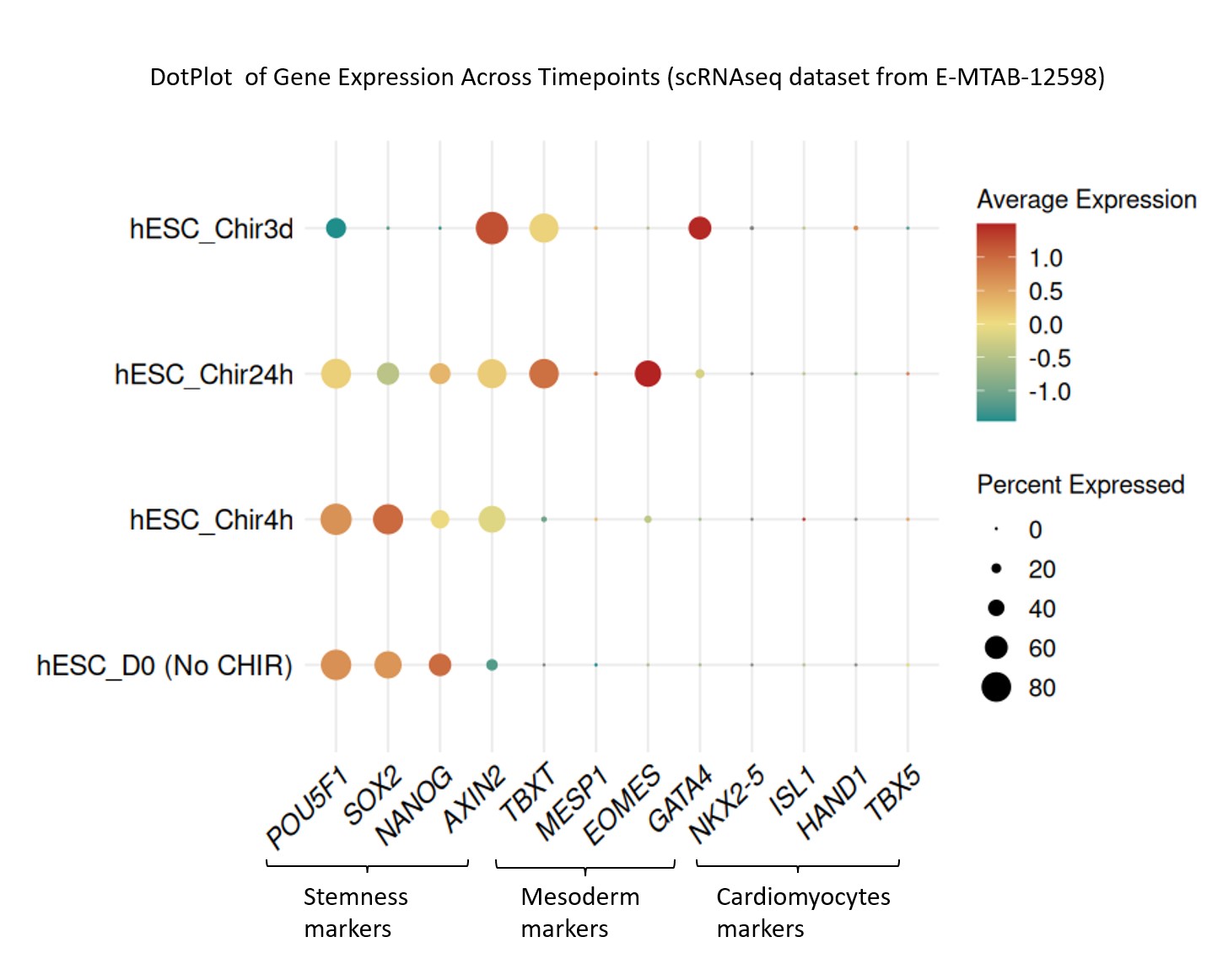
As my project is ongoing, I first performed a reanalysis of publicly available datasets (E-MTAB-12598) to establish a reference baseline. Using Seurat in R, I preprocessed scRNA-seq data, assessed batch effects via PCA, and applied Harmony for batch correction before UMAP visualization.
UMAP Results: The clustering shows a transition from pluripotency toward a mixed mesoderm-like state rather than a pure mesoderm population, likely due to CHIR-only treatment. scRNA-seq Analysis.
Dot Plot Trends: Stemness markers (POU5F1, SOX2) decrease over time, while mesoderm markers (TBXT, MESP1) increase. Cardiomyocyte markers (NKX2-5, ISL1) remain silent, confirming early differentiation but not definitive cardiomyocyte commitment.